2.1 Single FISH
2.1.1 RNA Probe Preparation
1.1.5 mL microcentrifuge tubes or standard 96-well V-bottom microplates (Greiner Bio-One, Longwood, FL, USA; Catalog No. 651201).
2.RNAse free DEPC-treated or double-distilled water.
3.T7, T3 or SP6 RNA Polymerase (Fermentas Life Sciences, Burlington, ON, Canada; Catalog Nos. EP0101, EP0111, EP0131) as appropriate.
4.T7, T3 or SP6 transcription buffer (supplied with RNA polymerases).
5.Fluorescein RNA labeling mix (Roche Applied Science, Laval, QC, Canada; Catalog No. 1 685 619).
6.Digoxigenin RNA labeling mix (Roche Applied Science; Catalog No. 1 277 073). For single labeling, choose item 5 or 6.
7.RNAguard (Amersham Biosciences, Piscataway, NJ, USA; Catalog No. 27-0816-01).
8.0.5 M EDTA.
9.4 M lithium chloride.
10.Absolute ethanol.
11.Cold 70% ethanol.
2.1.2 Initial Embryo Fixation
1.Chlorine bleach, diluted 1:1 with water.
2.40% formaldehyde solution (prepared fresh from paraformaldehyde as described in Subheading 3.1.2).
3.1x PBS solution (diluted from 10x PBS stock).
4.Heptane.
5.Methanol.
6.20 mL Disposable glass scintillation vials (Fisher Scientific Limited, Nepean, ON, Canada; Catalog No. 03-337-15).
2.1.3 Post-Fixation, Hybridization and Post-Hybridization Washes
1.1.5 and 0.5 mL microcentrifuge tubes, 0.2 mL 8-strip PCR tubes, or 0.2 mL 96-well PCR plates (Abgene, Rochester, NY, USA; Catalog No. AB-0900).
2.Microplate sealing film (Abgene; Catalog No. AB-1115).
3.PBT solution: 1x PBS plus 0.1% Tween-20.
4.40% formaldehyde solution, prepared that day (Subheading 3.1.2).
5.20 mg/mL (600 U/mL) proteinase K (Sigma Aldrich, Oakville, ON, Canada; Catalog No. P2308); dissolve in sterile water, divide into 10 μL aliquots and store at -20oC.
6.2 mg/mL glycine in PBS.
7.RNA hybridization solution: 50% formamide, 5x SSC, 100 μg/mL heparin, 100 μg/mL sonicated salmon sperm DNA and 0.1% Tween-20). Filter through a 0.2 micron filter and store at -20oC in aliquots (stable for at least 6-12 months).
8.Heating block(s) or water bath(s) adjustable to 56oC, 80oC, and 100oC.
2.1.4 Development of FISH Signal
1.1x PBS solution (diluted from 10x PBS stock).
2.PBT solution: 1x PBS, 0.1% Tween-20.
3.PBTB solution: 1x PBS, 0.1% Tween-20 and 1% milk powder.
4.TSA detection of digoxigenin:
a.Peroxidase-conjugated IgG fraction monoclonal mouse anti-digoxin (1/400 dilution of a 1 mg/mL stock solution; Jackson ImmunoResearch Laboratories Inc., West Grove PA, USA; Catalog No. 200-032-156) or
b.Biotin-SP-conjugated IgG fraction monoclonal mouse anti-digoxin (1/400 dilution of a 1 mg/ml stock solution; Jackson ImmunoResearch Laboratories Inc.; Catalog No. 200-062-156) and Streptavidin-HRP conjugate (1/100 dilution of a 1 μg/mL stock solution; Molecular Probes, Eugene OR, USA; Catalog No. S991). See Note 1 for advice on when to use a or b.
c.Alexa Fluor 488 tyramide conjugate (1/40 dilution of standard tyramide stock solution. Molecular Probes, Eugene OR, USA; Catalog No. T-20932) or Cyanine (Cy-3 or Cy5) tyramide conjugates (1/50 dilution of stock solution; Perkin Elmer Life Sciences, Boston, MA, USA; Catalog No. SAT704A or SAT705A).
5. Conventional fluorescence detection of fluorescein:
a.IgG fraction monoclonal mouse anti-fluorescein antibody (1/2000 dilution of a 0.1 mg/mL stock solution; Roche Applied Science; Catalog No. 1426 320).
b.Goat anti-mouse F(ab')2 fragment antibody conjugated to CY2 (1/2000 dilution of a 1 mg/mL stock solution; Jackson ImmunoResearch Laboratories Inc.; Catalog No. 115- 226-062).
2.1.5 Mounting and Viewing of Samples
1.Mountant: 70% glycerol, 2.5% DABCO (1,4-Diazabicyclo [2.2.2.] Octane; Sigma Aldrich; Catalog No. D-2522). In light-shielded tube, add 1.25 g of DABCO crystals to 15 mL of 1x PBS and rock until the DABCO dissolves. Add 35mL
of glycerol and shake vigorously for 2 to 3 min until glycerol dissolves. Store at - 20oC.
2.Microscope slides.
3.Coverslips (22x22 mm).
4.Fluorescence and/or LSC microscope.
2.2 Double FISH (see Notes 2 and 3)
Reagents for preparation of embryos and hybridization of labeled probes as described in Subheadings 2.1.3 and 2.1.4
1. Conventional detection of digoxigenin:
a. IgG fraction, sheep anti-digoxigenin antibody (1/1000 dilution of a 0.2 mg/ml stock solution; Roche Applied Science; Catalog No. 1333 089).
b. Donkey anti-sheep F(ab')2 fragments antibody conjugated to CY3 (1/2000 dilution of a 1mg/ml stock solution; Jackson ImmunoResearch Laboratories Inc.; Catalog No. 713- 166-147).
2. TSA detection of digoxigenin for double FISH:
a. Peroxidase-conjugated sheep anti-digoxigenin F(ab')2 fragments antibody (1/300 –
1/500 dilution of stock solution; Roche, Laval, QC, Canada; Catalog No. 1 207 733).
b. Tyramide Signal Amplification (TSA) Kits #3, #13, #23 with Alexa Fluor 546 tyramide (Molecular Probes, Eugene OR, USA; Catalog Nos. T-20913, T-20923 T- 20933).
3. Conventional detection of fluorescein: as described in Subheading 2.1.4 4. TSA detection of fluorescein for double FISH:
a. Peroxidase-conjugated rabbit anti-fluorescein antibody (1/400 – 1/600 dilution of stock solution; Molecular Probes, Eugene OR, USA; Catalog No. A-21253).
b. Tyramide Signal Amplification (TSA) Kits #2, #12 or #22, with Alexa Fluor 488 tyramide (Molecular Probes, Eugene OR, USA; Catalog Nos. T-20912, T-20922, T- 20932).
2.3 FISH on Dissected Tissues
1.1.5 mL microcentrifuge tubes.
2.1x PBS solution (diluted from 10x PBS stock).
3.40% formaldehyde solution (prepared fresh from paraformaldehyde as described in Subheading 3.1.2).
4.PBT solution: 1× PBS, 0.1% Tween-20.
5.Reagents for hybridization and detection of labeled probes as described in Subheadings 2.1.3 and 2.1.4.
2.4 RNA-Protein Double-labeling
1.Reagents for preparation of embryos, hybridization and detection of labeled probes as
described in Subheadings 2.1.3 and 2.1.4.
2.Primary antibody directed against the protein of interest. In order to prevent antibody cross-detection, make sure that the species origin of this antibody differs from that of the anti-digoxigenin or anti-fluorescein antibodies used to detect the FISH probe(s). In the example shown in Figure 2B-D, a rabbit polyclonal antibody against the Fushi tarazu (Ftz) homeodomain protein was utilized.
3.Select a fluorochrome-conjugated secondary antibody directed against the species of the primary antibody (see Note 3). In the case of the Ftz protein, a Cy3-conjugated Goat Anti-Rabbit IgG (H+L) (1/1000 dilution of a 1mg/mL stock solution; Jackson ImmunoResearch Laboratories Inc., West Grove. Catalog Nos. 111-165-144) was employed.
3. Methods
3.1 Single FISH
3.1.1 RNA Probe Preparation (see Note 4)
1. To prepare a run-off transcript from a plasmid template, linearize the plasmid to completion (see Note 5) with the appropriate restriction enzyme, and then remove the enzyme by successive phenol/chloroform and chloroform extractions. Heating to 65°C for 15 min after the last chloroform extraction helps to remove residual chloroform. To precipitate the DNA, add NaAcetate (pH 5.2) to 0.3 M followed by three volumes of ethanol, then cool to -70°C for at least 20 min. Spin 10 min in a cold microcentrifuge at 13,000 rpm and wash with cold 70% ethanol. Alternatively, the linearized template can be purified following agarose gel electrophoresis using common gel extraction kits, such as the Minelute Gel Extraction Kit from Qiagen Inc. (Mississauga, ON, Canada; Catalog No. 28604), according to the manufacturers’ recommendations. As a precaution against RNase contamination, the gel extraction kit should be reserved for the preparation of transcription templates, and gloves should be used when handling components of the kit. We generally prepare 5-10 μg of linearized template, resuspended or eluted in 20-40 μL RNAse-free water.
Alternatively, a PCR product can be used as the template for in vitro transcription. If a PCR-amplified template is large (>3kb), the products of the in vitro transcription reaction will be of variable length, but this does not impair their effectiveness as probes. This method is particularly convenient when transcripts for many genes are being prepared simultaneously. Use PCR to amplify a DNA fragment containing the gene of interest and flanking promoters (T3, T7, or Sp6). For most genes in the Drosophila genome, cDNA sequences cloned between flanking promoters are readily available from cDNA libraries; PCR protocols for the vectors used in these libraries are described by Tomancak et al. (2002) (7). The PCR products can be purified and precipitated as described above. For templates that are amplified in a 96-well plate format, the PCR products can be bulk purified using special filtration plates (Whatman Inc.; Clifton, NJ, USA; Catalog No. 7700-1303), according to the manufacturers’ recommendations. Purified products can then be concentrated by ethanol precipitation in standard V-bottom 96-well plates, using a centrifuge rotor with microplate adaptors, and then resuspended in 10 μL of RNAse-free water.
2.RNA probes are prepared as described by Roche Applied Science, on the product sheets of their digoxigenin and fluorescein RNA labeling kits. On ice, add 0.5-1 μg linearized template DNA or PCR product, 2 μL fluorescein or digoxigenin RNA labeling mix, 2 μL 10x transcription buffer (supplied with the RNA polymerase: 0.4 M Tris-HCl, pH 8.0; 60 mM MgCl2, 100 mM dithiothreitol, 20 mM spermidine), 1 μL RNAguard (40 U/μL) and sterile, RNAse-free water to make the final reaction volume equal to 18 μL. Add 2 μL of the appropriate RNA polymerase (T3/T7/SP6; 20 U/μL), mix well and incubate at 37°C for 2 h.
For PCR templates amplified and purified in 96-well format, probes can be bulk synthesized in V-bottom microplates in a total reaction volume of 10 μl. In each well, 5 μL of resuspended template is combined with 5 μL of pre-mixed and pre-aliquoted transcription reaction mixture containing: 1 μL 10x transcription buffer, 0.5 μL digoxigenin labeling mix, 0.4 μL RNA polymerase (20 U/μL), 0.125 μL RNAguard (40 U/μL), and 3 μL RNAse-free water. Plates are then sealed with adhesive foil and incubated for 2 h at 37°C.
3.Once probe synthesis is completed, reactions are stopped with EDTA (20 mM final concentration), and the probes are precipitated by adding LiCl (50 mM final concentration), and 2.5 volumes of absolute ethanol (see Note 6). Chill to -70°C, spin and wash the pellets as described above. After drying, resuspend the pellets in 50-100 μL RNAse-free water (see Note 7). Check the probes by loading and running 5 μL on a conventional agarose gel (~1%). The run-off transcript(s) should easily be detected by ethidium bromide staining (4). Estimate the concentration of the probe solution by comparison with a conventional DNA ladder; this estimate will be used to determine the amount of probe to add to the in situ reactions. Probes should be stored at -20°C. Several freeze/thaw cycles on ice do not impair probe activity.
3.1.2 Initial Embryo Fixation
-
1.Prepare the 40% formaldehyde stock solution just prior to embryo dechorionation (see Note 8). Dissolve 0.92 g paraformaldehyde in 2.5 mL water containing 35 μL 1N KOH. Heat the mixture at 80°C until the paraformaldehyde dissolves and then store it at 4°C until ready for use. At that time, filter the solution through a 0.45 micron filter.
-
2.Collect and rinse embryos in water.
-
3.Dechorionate the collected embryos in a 1:1 mixture of chlorine bleach and water for approximately 90 s. When dechorionated, the embryos will either float to the surface of the bleach solution or stick to the sides of the collection basket. Embryos should be rinsed immediately as over-dechorionation is detrimental. Rinse the collection basket with plenty of water. Fast flowing tap water helps to dechorionate partially dechorionated embryos. An optional rinse with 0.7% NaCl, 0.03% Triton X-100 is helpful for removing residual bleach and for washing embryos down from the sides of the basket.
-
4.Transfer the embryos to a 20 mL glass scintillation vial (see Note 9) containing a biphasic mixture of 8 mL heptane, 2.5 mL 1x PBS and 250 μL 40% formaldehyde. Shake for 20 min (embryos need alternating exposure to both phases for uniform permeabilization and fixation; gentle intermittent swirling is usually sufficient).
-
5.Using a 1 mL pipetteman, draw up embryos (which are at the interphase), taking care not to suck up any of the lower aqueous phase (see Note 10). Transfer to a 1.5 mL microfuge tube containing 0.5 mL heptane and 0.5 mL methanol for devitellinization. Shake vigorously until most of the embryos sink to the bottom (about 30 sec). Carefully remove about 75% of the heptane and methanol and replace with 1 mL methanol. Shake once more. All or most of the embryos should now sink to the bottom of the tube. Remove all of the liquid along with any embryos remaining at the interphase, then rinse 2 to 3 times with methanol. Embryos can be stored in methanol at -20°C for several months.
3.1.3 Post-Fixation, Hybridization and Post-Hybridization Washes
The following steps are optimized for fixations performed in 1.5 mL microfuge tubes (50 μL settled embryos), 0.2 mL 8-well PCR strips or 0.2 mL 96-well PCR plates (10 μL settled embryos/well). The latter are particularly well suited for the use of multiple probes, probe combinations or when testing other conditions. Unless otherwise indicated, the wash volumes used below are 1 mL or 175 μL for 1.5 mL tubes or 0.2 mL PCR tubes, respectively. If not, the appropriate volumes for each tube format are provided (separated by or as above).
1.Aliquot embryos in microfuge tubes, PCR strips, or PCR plates using wide aperture tips (cut ends off as necessary). Rinse the embryos once in methanol.
2.Rinse the embryos 2 times in PBT (1x PBS, 0.1% Tween-20).
3.Post-fix the embryos for 20 min in 4% formaldehyde (prepared by diluting fresh 40% formaldehyde 1/10 in PBT). Place tubes on a rocking platform or rotating mixer to ensure even fixation. If using PCR plates, secure plate in a vertical position to achieve more efficient mixing.
4.Wash embryos 3 times in PBT for approximately 2 minutes/wash.
5.Dilute the 20 mg/mL (600 U/mL) stock proteinase K solution in PBT to a final concentration of 3 μg/mL (0.09 U/mL) (see Note 11). Add 500 μL or 100 μL of 3 μg/mL proteinase K to each sample of embryos and incubate at room temperature for either 2 or 13 min, depending on age (see Note 12). During this period, mix 2 to 3 times by drawing up some of the solution with a pipetteman and jetting it back into the tubes/wells such that the embryos are gently mixed back into suspension. At the end of the incubation at room temperature, transfer the embryos to ice and incubate for an additional hour. This reduction in proteinase K concentration and increase in incubation time was found to be critical for obtaining optimal uniform and consistent detection of all RNAs.
6.Remove proteinase K solution and block digestion by adding PBT containing 2 mg/mL glycine. Remove after 2 min and wash for another 2 min with the same solution.
7.Rinse embryos 2 times in PBT to remove the glycine.
8.Post-fix the embryos again (as in step 3) for 20 minutes in PBT containing 4% formaldehyde.
9.Wash embryos 5 times in PBT, 5 min/wash, to remove all traces of fixative
10.Rinse the embryos in a 1:1 mixture of PBT and RNA hybridization solution. Replace the mixture with 100% hybridization solution. At this point, the embryos can be stored overnight at -20°C.
11.In a separate tube, boil 500 μL or 100 μL of RNA hybridization solution at 100°C for 5 min, then cool it on ice for 5 min. This freshly boiled hybridization solution will be used as the pre-hybridization solution.
12.If using 1.5 mL microfuge tubes to this point, transfer to 0.5 mL tubes, remove the hybridization solution from the tube/wells and place on ice. Add the freshly boiled and cooled pre-hybridization solution and place the embryos in a 56°C heat block/water bath. If using microtiter plates, cover and seal. Incubate at 56°C for a minimum of 2 h.
13.After the pre-hybridization, remove the pre-hybridization solution and add 100 μL hybridization solution plus probe. Though optimal probe concentration may have to be determined empirically, 50 ng of probe in 100 μL of RNA hybridization solution usually works well when followed by TSA. If conventional alkaline phosphatase or fluorescently-conjugated antibodies are going to be used, about 200 ng of probe should be added. The probe diluted in hybridization buffer is heated to 80°C for 3 min, cooled on ice for 5 min and then added to the embryos. Extremely short probes (<300 bp) should be heated at 80°C for only 1 min.
14.Hybridizations are carried out at 56°C for 12 to 16 h.
15.Pre-heat all wash solutions to 56°C. Remove the probe solution and rinse the embryos once with 400 μL or 175 μL pre-warmed hybridization buffer. Replace the rinse solution with another 400 μL or 175 μL pre-warmed hybridization buffer and incubate at 56°C for 20-30 min.
16.Wash for 15 min each with 400 μL or 175 μL of 3:1, 1:1 and 1:3 mixtures of hybridization buffer and PBT.
17.Wash 4 times, 5 min/wash, with 400 μL or 175 μL pre-warmed PBT, then cool embryos to room temperature.
3.1.4 Development of FISH Signal
Unless otherwise indicated, the wash volumes used below are 400 μL or 175 μL for 0.5 mL tubes or 0.2 mL PCR strips/plates, respectively.
1.Add PBTB (1x PBS, 0.1% Tween-20, 1.0% milk powder) and incubate for 10 min with constant mixing. The use of milk powder in this and subsequent steps helps to reduce background (see Note 13).
2.The first step in detecting hybridized RNA probes is to incubate the embryos with the appropriate primary antibodies diluted in PBTB. For single FISH, we typically use a DIG-labeled probe and detection by tyramide signal amplification for high sensitivity (see Notes 1 and 2). The dilutions that were optimal in our hands are given in Subheading 2.1.4; however, due to variations in the reagents, equipment and methodology used by different labs, the dilution for optimal activity should be determined empirically. Incubate with primary antibodies for 2 h with constant mixing.
3.Wash for 2 h with 5 changes of PBTB. If an HRP-conjugated primary antibody is being used for TSA detection, proceed directly to Step 6.
4.Add the appropriate secondary antibody(ies) diluted in PBTB, and incubate with constant mixing for 2 h. If streptavidin-HRP is being used as a secondary detection reagent for TSA, 1 h of incubation is sufficient. For fluorescently-conjugated secondary antibodies, perform this step and all subsequent steps in light-shielded tubes (e.g. cover tubes with aluminum foil or work with reduced light).
5.Wash for 2 h with 5 changes of PBTB, then wash 2 times with PBT.
6.If TSA is being used, carry out the TSA reaction as follows (this protocol was adapted from the TSA kit product sheets). Wash the embryos 3 times in 1x PBS for 5 min/wash. Prepare dilutions of the appropriate tyramide substrate in amplification buffer provided with the substrate (see Note 14). Remove the last PBS wash from the embryos, add 100 μL or 40 μL tyramide solution, and incubate in the dark at room temperature for 2 h with constant mixing. After the reaction, rinse the embryos in PBS, then wash them for 2 h with 5 changes of PBS.
3.1.5 Mounting and Viewing of Samples
1.Resuspend embryos in DABCO-containing mountant. Allow the embryos to settle to the bottom of the tube (1-3 h or overnight at 4°C) before mounting.
2.Transfer the embryos to a clean slide in ~35 μL mountant and cover with a 22x22 mm coverslip. Seal the edges with nail polish. Slides can be stored for weeks at 4°C in the dark. Background levels will often decrease over the first few days.
3.Embryos can be viewed by either conventional fluorescence microscopy, LSCM or deconvolution microscopy.
3.2 Double Fluorescent in situ Hybridization
1.Generate two probes, each with a different label, as described in Subheading 3.1.1
(see Note 4 for alternative/additional labels).
2.Collect and fix embryos as described in Subheading 3.1.2.
3.Perform the hybridization with both probes simultaneously; all other pre- and post- hybridization washes are as described in Subheading 3.1.3.
4.Add both anti-digoxigenin and anti-fluorescein primary antibodies (see Note 3); incubate and wash as described in Subheading 3.1.4.
5.Detect the primary antibodies using the appropriate secondary detection reagents (see Note 3). If conventional fluorescently-conjugated secondary antibodies are being used for both probes, add them simultaneously. If one of the probes is being detected by TSA, add the HRP-conjugate along with the fluorescently-conjugated secondary antibody, then carry out the TSA reaction as described for single FISH. If both probes are being detected by TSA, add the first HRP-conjugate, perform the TSA reaction, and quench as described in Note 15 before adding the second HRP-conjugate and performing the second reaction. Regardless of detection technique, carry out the first addition of fluorochrome and all subsequent steps in light shielded tubes.
6.Mount samples as described in Subheading 3.1.5.
3.3 FISH on Dissected Tissues
1.Dissect tissues, such as imaginal disks and salivary glands, in 1× PBS. Dissected tissues can be stored briefly (up to 30 min) on ice in a microfuge tube containing PBS until enough tissue for analysis is obtained.
2.Remove the PBS and add 600 μL of fixation solution (540 μL 1× PBS and 60 μL 40% formaldehyde). Shake gently for 20 min. (see Note 16)
4.Rinse the tissues in 1 mL of 1:1 mixture of PBT and RNA hybridization solution. Replace the mixture with 100% hybridization solution and pre-hybridize the embryos at 56oC for a minimum of 2 h.
5.After pre-hybridization, remove hybridization solution and add 100 μL of hybridization solution containing the probe. Optimal probe concentration needs to be determined empirically but generally 50 ng probe in 100 μL of RNA hybridization solution works well if using TSA. Diluted probe is heated to 100oC for 5 min, cooled for 5 min on ice and then added to the tissues.
6.Perform hybridization, post-hybridization washes, antibody incubations, and TSA reaction as described in Subheading 3.1.4.
7.Mount samples as described in Subheading 3.1.5.
Figure 2A shows an example Drosophila brain tissue staining for dE75 mRNA.
3.4 RNA-Protein Double-labeling
1.Collect and fix embryos as described in Subheading 3.1.2, with the exception of the proteinase K step. The proteinase K concentration may have to be lowered further to preserve integrity of protein epitopes (see Note 11).
2.Perform the hybridization and washes, as described in Subheading 3.1.3.
3.Add the primary antibody against the protein of interest, along with the anti- fluorescein or anti-digoxygenin antibody to detect the FISH probe(s), and then incubate and wash as in Subheading 3.1.4. To obtain differential signals, both primary antibodies must have been raised in different hosts.
4.Add both secondary detection reagents (fluorochrome-conjugated secondary antibodies and streptavidin-HRP) (see Note 3). Perform incubations, washes, and TSA reaction as in Subheading 3.1.4.
5.Mount samples as described in Subheading 3.1.5.
Figure 2B-D shows an example of co-staining of the Ftz mRNA and protein in cellular
blastoderm embryos (note incomplete overlap).
4. Notes
1.The peroxidase-conjugated monoclonal mouse anti-digoxin is ideal for use in single FISH with TSA because it gives a strong signal and has very low background. The biotin-SP-conjugated monoclonal mouse anti-digoxin, when used in conjunction with HRP-streptavidin from the TSA kit, gives much stronger tyramide signal amplification than the peroxidase-conjugated anti-digoxin alone. This is useful for amplifying extremely weak signals, but is unnecessary for moderate to strong signals and adds additional steps to an already lengthy protocol.
2.We have not yet tested TSA reagents for the detection of fluorescein. However, Kosman et al. (2004) (8) have provided a detailed description of the reagents used for the detection of fluorescein labeled probes by TSA. These may give enhanced sensitivity of detection if substituted for the conventional fluorescence detection reagents that we have used. Kosman et al. suggest that these antibodies do not work well with milk or BSA as blocking reagents, and have recommended the Western Blocking Reagent (Catalog No. 1 921 673) from Roche Applied Science as a substitute (8). However, we have used the peroxidase-conjugated sheep anti- digoxigenin F(ab')2 fragments antibody in single FISH, and have not had a problem using milk as a blocker.
3.The antibodies described here for conventional fluorescence detection have been selected for usefulness in double-labeling. The primary antibodies are whole IgGs raised in different hosts. Similarly, secondary antibodies are selected so that they are unlikely to cross react with the second primary antibody or with each other. Jackson Immunoresearch, from whom the recommended secondary antibodies were obtained, provides information, suggestions and many products that make choosing and obtaining the appropriate antibodies relatively easy. Secondary antibodies most suitable for multiple-labeling are designated "ML". These are generally comprised of the F(ab')2 portion of IgG antibodies that recognize both heavy and light (H and L) chains of their target antibodies. ML antibodies are also preadsorbed against multiple host sera. For this reason, and because the antibodies contain light sensitive molecules, we do not bother to preadsorb them against embryos. However, if background is high, this may help.
4.The digoxigenin and fluorescein labels described here can be substituted for, or supplemented with, other labels. Examples include biotin, dinitrophenyl (DNP) and a number of new Alexa-conjugated nucleotides. The latter are detected by Alexa- specific (fluorescently-conjugated if desired) antibodies, such as those directed against Alexa Fluor 405 and Alexa Fluor 488 (Molecular Probes; Catalog No. A-5760 and A-11094). These reagents provide numerous possibilities for multiple RNA pattern recognition. Kosman et al. (2004) describe the detection of three or more labels (8).
5.Template DNA should be chosen and linearized such that run-off transcripts correspond to unique portions of the gene's coding region. So far, we've found that run-off transcripts ranging from about 0.4-1 kb work well as probes. Cutting to completion generally takes 2-4 h, and should be confirmed by agarose gel electrophoresis.
6.Removal of template with DNAseI subsequent to the transcription reaction is unnecessary. Precipitation of the probe with LiCl removes most unincorporated nucleotides.
7.Some in situ protocols use carbonate degradation to reduce the size of their RNA transcripts for increased embryo penetration. In our hands this was found to be unnecessary and even detrimental.
8.Freshly prepared formaldehyde is a stronger fixative than commercially prepared formaldehyde solutions. A strong fixation with fresh formaldehyde is essential for maintaining embryo integrity and morphology during hybridization and following proteinase K digestion.
9.Vessel sizes used here are optimized for small collections (<250 μL settled embryos). For larger sample sizes, larger vessels should be used, keeping the same relative ratios. 50 mL polypropylene Falcon tubes work well for fixing and devitillinizing settled embryo volumes from ~0.25 to 2 mL. Care should be used as some tubes or plastics appear to interfere with fixation and devitellinisation (Sarstedt polystyrene tubes for example).
10.If transferred to the devitellinization solution along with the embryos, the aqueous portion of the fixation mix will impair devitellinization. This has likely occurred if the devitellinization solution is cloudy and less than 80% of embryos move from the interphase to the bottom of the tube. Care should be taken to minimize uptake of the lower aqueous phase when drawing up embryos from the fixative into the pipette tip. If this occurs, the phases often separate in the tip, and the lower aqueous phase can be returned to the scintillation vial. If transfer of aqueous solution has already occurred, the devitellinization step can be repeated as necessary by removing as much heptane and methanol as possible, replacing with fresh heptane and methanol and shaking again.
11.Optimal proteinase K digestion is critical for probe entry into the embryo. Too little, and the resulting signal is very weak. Also, tissues deeper within the embryo are particularly dependent on proteinase treatment. On the other hand, over-digestion results in poor embryo integrity and morphology. During RNA-protein double- labeling, this also leads to the destruction of protein epitopes. Previous protocols call for a 5 min incubation at room temperature with high concentrations of proteinase K. We find that this short incubation time led to excessive degradation at the embryo surface and variable digestion within. There also tends to be a great deal of variation from embryo to embryo and sample to sample. We have remedied these problems by lowering the Proteinase K concentration, increasing the incubation time and reducing the temperature. With this protocol, we obtain increased probe sensitivity, penetration and uniformity from embryo to embryo.
12.We recommend a 2 min incubation at room temperature, followed by 60 min on ice, for embryos older than 4 h AEL. Prior to 4 h AEL, the duration of the room temperature incubation should be increased to 13 min (the incubation on ice remains unchanged).
13.The concentration of milk used in this protocol (i.e. 1%) has been optimized for use with the peroxidase- and biotin-SP-conjugated anti-digoxin antibodies described in Subheading 2.1.4. For other antibodies, the concentration of milk can be varied as necessary to increase the signal and reduce the background. Weak signals can sometimes be strengthened by decreasing the milk concentration to 0.5%. Conversely, high background levels can be reduced by raising the milk concentration; some antibodies with poor specificity work best with milk concentrations as high as 4%.
14.If using Alexa-tyramide conjugates from Molecular Probes, dilute the 30% hydrogen peroxide in kit amplification buffer to a final concentration of 0.0015%. Dilute the tyramide substrate 1/40 in this solution. If using Cyanine-tyramide conjugates from Perkin Elmer Life Sciences, directly dilute the tyramide substrate 1/50 in the amplification buffer provided with the kit, without adding hydrogen peroxide. 1μL of substrate in a 100 μL reaction is standard, but more tyramide can be added if a stronger signal is desired, and 2.5 μL has been found to be saturating in our hands.
15. Three methods have been suggested for quenching HRP. Kosman et al. (2004) recommend incubating the embryos in PBT + 1% hydrogen peroxide for 20 minutes with constant mixing; the hydrogen peroxide is then removed by three rinses in PBT, followed by a 5 min wash in PBT with constant mixing. Wilkie and Davis (1998) recommend a 10 min incubation in 0.01 M HCl (strips off Ab), followed by washes to remove the HCl. Alternatively, they suggest incubating the embryos at 70°C for 15 min.
16. Dissected larval tissues tend to be more sensitive to proteinase K digestion than embryos; as a result, we often omit the proteinase K step when dealing with such samples, and have not rigorously optimized the digestion conditions for them. We have tried a 45 s incubation at room temperature with 2.14 U/ml proteinase K (with no further incubation on ice), and this has worked adequately.

FISH protocols for Drosophila

These are some of the methods we use regularly in the lab and that we thought might prove useful to others in the field. Please feel free to send comments, corrections or improvements. If you're looking for other Drosophila based methods, a good place to start is here.
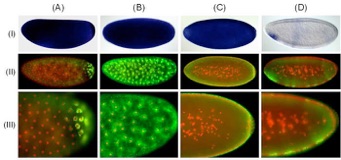
Fig. 1. Comparison of alkaline phosphatase and tyramide-amplified detection of mRNAs. Staining patterns obtained using DIG-labeled antisense probes directed against the (A) CG14217, (B,C) Bicaudal-D, and (D) Charybde mRNAs, through conventional AP-based detection (I) or tyramide signal amplification (II), using tyramide-Alexa Fluor 488 (green fluorescence). Close-up images of tyramide-amplified samples are also shown (III). (A) Transcripts of the CG14217 gene demonstrate posterior localization in ring-shaped structures that surround the blastoderm nuclei. (B-C) Bicaudal-D transcripts exhibit highly dynamic localization patterns during Drosophila embryogenesis, including (B) enrichment in perinuclear structures surrounding migrating nuclei and (C) apical localization above peripherally located blastoderm nuclei. (D) During cellularization,Charybde mRNA is detected in nuclear foci representing nascent zygotic transcripts. In rows II and III, nuclei were labeled in red with propidium iodide.
Fig. 2. Examples of dissected tissue staining and RNA-protein co-detection. (A) Hybridization performed on Drosophila brain tissue dissected at the pre-pupal stage; using a DIG-labeled antisense probe fordE75 mRNA amplified using Tyramide-Alexa Fluor 488 (green fluorescence. Nuclei were counter-stained in red with propidium iodide. (B-D) Co-detection of fushi tarazu (ftz) mRNA and protein. (B) Embryo hybridized with a DIG-labeled antisense probe for ftz mRNA detected using Tyramide-Alexa Fluor 488 (green fluorescence). (C) Detection of the Ftz protein was accomplished using a polyclonal antibody raised against Ftz and a Cy3-conjugated secondary antibody (red fluorescence). (D) Superimposed images of ftz RNA and protein. Nuclei were labeled in blue with 4'-6-Diamidino-2-phenylindole (DAPI).